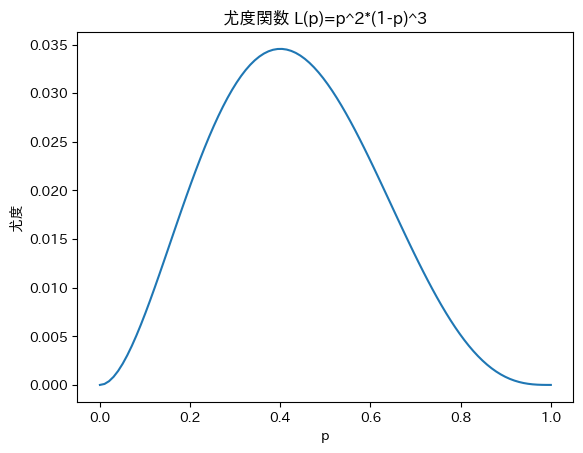
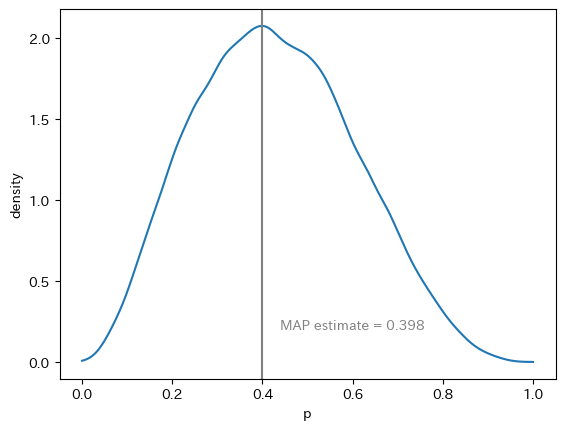
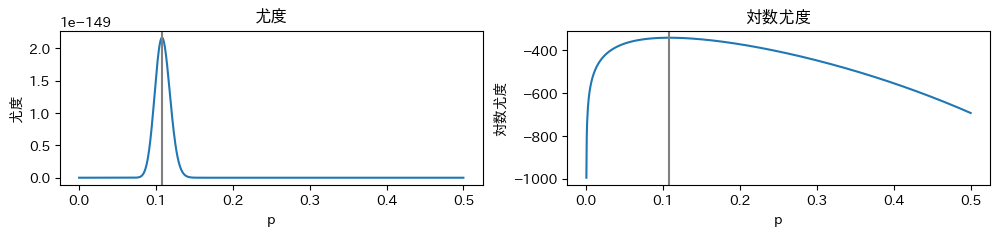
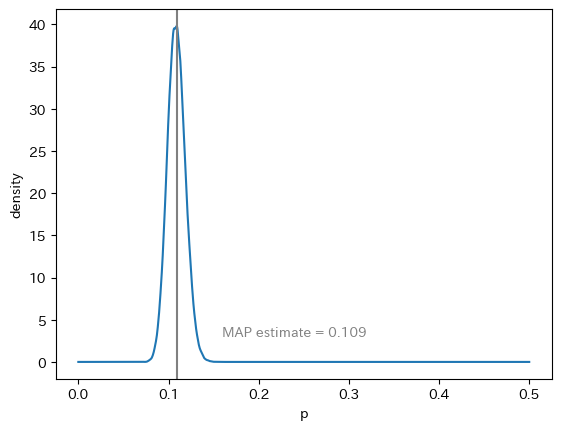
In file included from /usr/local/lib/python3.9/site-packages/httpstan/include/boost/multi_array/multi_array_ref.hpp:32,
from /usr/local/lib/python3.9/site-packages/httpstan/include/boost/multi_array.hpp:34,
from /usr/local/lib/python3.9/site-packages/httpstan/include/boost/numeric/odeint/algebra/multi_array_algebra.hpp:22,
from /usr/local/lib/python3.9/site-packages/httpstan/include/boost/numeric/odeint.hpp:63,
from /usr/local/lib/python3.9/site-packages/httpstan/include/stan/math/prim/functor/ode_rk45.hpp:9,
from /usr/local/lib/python3.9/site-packages/httpstan/include/stan/math/prim/functor/integrate_ode_rk45.hpp:6,
from /usr/local/lib/python3.9/site-packages/httpstan/include/stan/math/prim/functor.hpp:14,
from /usr/local/lib/python3.9/site-packages/httpstan/include/stan/math/rev/fun.hpp:196,
from /usr/local/lib/python3.9/site-packages/httpstan/include/stan/math/rev.hpp:10,
from /usr/local/lib/python3.9/site-packages/httpstan/include/stan/math.hpp:19,
from /usr/local/lib/python3.9/site-packages/httpstan/include/stan/model/model_header.hpp:4,
from /root/.cache/httpstan/4.8.2/models/5zjucfky/model_5zjucfky.cpp:2:
/usr/local/lib/python3.9/site-packages/httpstan/include/boost/functional.hpp:180:45: warning: ‘template<class _Arg, class _Result> struct std::unary_function’ is deprecated [-Wdeprecated-declarations]
180 | : public boost::functional::detail::unary_function<typename unary_traits<Predicate>::argument_type,bool>
| ^~~~~~~~~~~~~~
In file included from /usr/include/c++/12/string:48,
from /usr/include/c++/12/bits/locale_classes.h:40,
from /usr/include/c++/12/bits/ios_base.h:41,
from /usr/include/c++/12/ios:42,
from /usr/include/c++/12/istream:38,
from /usr/include/c++/12/sstream:38,
from /usr/include/c++/12/complex:45,
from /usr/local/lib/python3.9/site-packages/httpstan/include/Eigen/Core:96,
from /usr/local/lib/python3.9/site-packages/httpstan/include/Eigen/Dense:1,
from /usr/local/lib/python3.9/site-packages/httpstan/include/stan/math/prim/fun/Eigen.hpp:22,
from /usr/local/lib/python3.9/site-packages/httpstan/include/stan/math/rev.hpp:4:
/usr/include/c++/12/bits/stl_function.h:117:12: note: declared here
117 | struct unary_function
| ^~~~~~~~~~~~~~
/usr/local/lib/python3.9/site-packages/httpstan/include/boost/functional.hpp:214:45: warning: ‘template<class _Arg1, class _Arg2, class _Result> struct std::binary_function’ is deprecated [-Wdeprecated-declarations]
214 | : public boost::functional::detail::binary_function<
| ^~~~~~~~~~~~~~~
/usr/include/c++/12/bits/stl_function.h:131:12: note: declared here
131 | struct binary_function
| ^~~~~~~~~~~~~~~
/usr/local/lib/python3.9/site-packages/httpstan/include/boost/functional.hpp:252:45: warning: ‘template<class _Arg, class _Result> struct std::unary_function’ is deprecated [-Wdeprecated-declarations]
252 | : public boost::functional::detail::unary_function<
| ^~~~~~~~~~~~~~
/usr/include/c++/12/bits/stl_function.h:117:12: note: declared here
117 | struct unary_function
| ^~~~~~~~~~~~~~
/usr/local/lib/python3.9/site-packages/httpstan/include/boost/functional.hpp:299:45: warning: ‘template<class _Arg, class _Result> struct std::unary_function’ is deprecated [-Wdeprecated-declarations]
299 | : public boost::functional::detail::unary_function<
| ^~~~~~~~~~~~~~
/usr/include/c++/12/bits/stl_function.h:117:12: note: declared here
117 | struct unary_function
| ^~~~~~~~~~~~~~
/usr/local/lib/python3.9/site-packages/httpstan/include/boost/functional.hpp:345:57: warning: ‘template<class _Arg, class _Result> struct std::unary_function’ is deprecated [-Wdeprecated-declarations]
345 | class mem_fun_t : public boost::functional::detail::unary_function<T*, S>
| ^~~~~~~~~~~~~~
/usr/include/c++/12/bits/stl_function.h:117:12: note: declared here
117 | struct unary_function
| ^~~~~~~~~~~~~~
/usr/local/lib/python3.9/site-packages/httpstan/include/boost/functional.hpp:361:58: warning: ‘template<class _Arg1, class _Arg2, class _Result> struct std::binary_function’ is deprecated [-Wdeprecated-declarations]
361 | class mem_fun1_t : public boost::functional::detail::binary_function<T*, A, S>
| ^~~~~~~~~~~~~~~
/usr/include/c++/12/bits/stl_function.h:131:12: note: declared here
131 | struct binary_function
| ^~~~~~~~~~~~~~~
/usr/local/lib/python3.9/site-packages/httpstan/include/boost/functional.hpp:377:63: warning: ‘template<class _Arg, class _Result> struct std::unary_function’ is deprecated [-Wdeprecated-declarations]
377 | class const_mem_fun_t : public boost::functional::detail::unary_function<const T*, S>
| ^~~~~~~~~~~~~~
/usr/include/c++/12/bits/stl_function.h:117:12: note: declared here
117 | struct unary_function
| ^~~~~~~~~~~~~~
/usr/local/lib/python3.9/site-packages/httpstan/include/boost/functional.hpp:393:64: warning: ‘template<class _Arg1, class _Arg2, class _Result> struct std::binary_function’ is deprecated [-Wdeprecated-declarations]
393 | class const_mem_fun1_t : public boost::functional::detail::binary_function<const T*, A, S>
| ^~~~~~~~~~~~~~~
/usr/include/c++/12/bits/stl_function.h:131:12: note: declared here
131 | struct binary_function
| ^~~~~~~~~~~~~~~
/usr/local/lib/python3.9/site-packages/httpstan/include/boost/functional.hpp:438:61: warning: ‘template<class _Arg, class _Result> struct std::unary_function’ is deprecated [-Wdeprecated-declarations]
438 | class mem_fun_ref_t : public boost::functional::detail::unary_function<T&, S>
| ^~~~~~~~~~~~~~
/usr/include/c++/12/bits/stl_function.h:117:12: note: declared here
117 | struct unary_function
| ^~~~~~~~~~~~~~
/usr/local/lib/python3.9/site-packages/httpstan/include/boost/functional.hpp:454:62: warning: ‘template<class _Arg1, class _Arg2, class _Result> struct std::binary_function’ is deprecated [-Wdeprecated-declarations]
454 | class mem_fun1_ref_t : public boost::functional::detail::binary_function<T&, A, S>
| ^~~~~~~~~~~~~~~
/usr/include/c++/12/bits/stl_function.h:131:12: note: declared here
131 | struct binary_function
| ^~~~~~~~~~~~~~~
/usr/local/lib/python3.9/site-packages/httpstan/include/boost/functional.hpp:470:67: warning: ‘template<class _Arg, class _Result> struct std::unary_function’ is deprecated [-Wdeprecated-declarations]
470 | class const_mem_fun_ref_t : public boost::functional::detail::unary_function<const T&, S>
| ^~~~~~~~~~~~~~
/usr/include/c++/12/bits/stl_function.h:117:12: note: declared here
117 | struct unary_function
| ^~~~~~~~~~~~~~
/usr/local/lib/python3.9/site-packages/httpstan/include/boost/functional.hpp:487:68: warning: ‘template<class _Arg1, class _Arg2, class _Result> struct std::binary_function’ is deprecated [-Wdeprecated-declarations]
487 | class const_mem_fun1_ref_t : public boost::functional::detail::binary_function<const T&, A, S>
| ^~~~~~~~~~~~~~~
/usr/include/c++/12/bits/stl_function.h:131:12: note: declared here
131 | struct binary_function
| ^~~~~~~~~~~~~~~
/usr/local/lib/python3.9/site-packages/httpstan/include/boost/functional.hpp:533:73: warning: ‘template<class _Arg, class _Result> struct std::unary_function’ is deprecated [-Wdeprecated-declarations]
533 | class pointer_to_unary_function : public boost::functional::detail::unary_function<Arg,Result>
| ^~~~~~~~~~~~~~
/usr/include/c++/12/bits/stl_function.h:117:12: note: declared here
117 | struct unary_function
| ^~~~~~~~~~~~~~
/usr/local/lib/python3.9/site-packages/httpstan/include/boost/functional.hpp:557:74: warning: ‘template<class _Arg1, class _Arg2, class _Result> struct std::binary_function’ is deprecated [-Wdeprecated-declarations]
557 | class pointer_to_binary_function : public boost::functional::detail::binary_function<Arg1,Arg2,Result>
| ^~~~~~~~~~~~~~~
/usr/include/c++/12/bits/stl_function.h:131:12: note: declared here
131 | struct binary_function
| ^~~~~~~~~~~~~~~